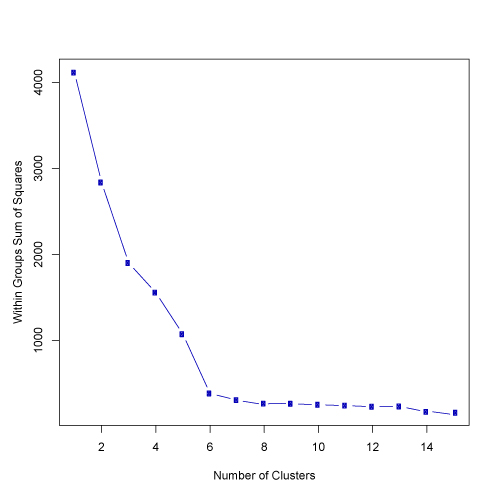
click to see image full size
In many cases, however, there will not be such an obvious break in the distribution of SSE against cluster solutions. To help in such cases, the kmeans.R script conducts additional analyses to evaluate cluster solutions. Specifically, the script produces 250 randomized versions of the original input data, and calculates SSE against cluster solutions for the randomized data. The data is randomized by column, so each variable will have the same mean and standard deviation in both the actual and randomized matrices. If a data set has strong clusters, the SSE of the actual data should decrease more quickly than the random data as than cluster level goes up. Thus, the kmeans.R script plots SSE against the number of tested clusters for both the actual and 250 randomized matrices. Plots below are shown on both a log scale (left) and on a normal scale (right).
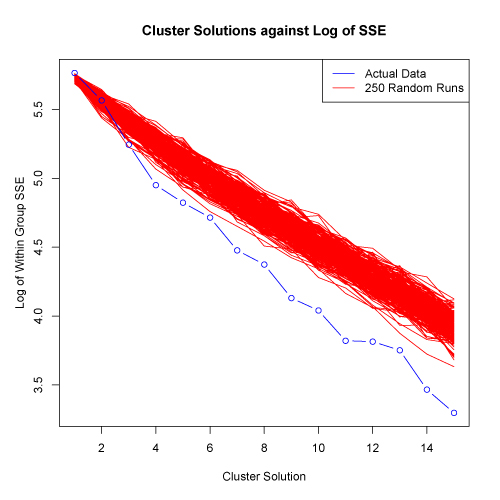
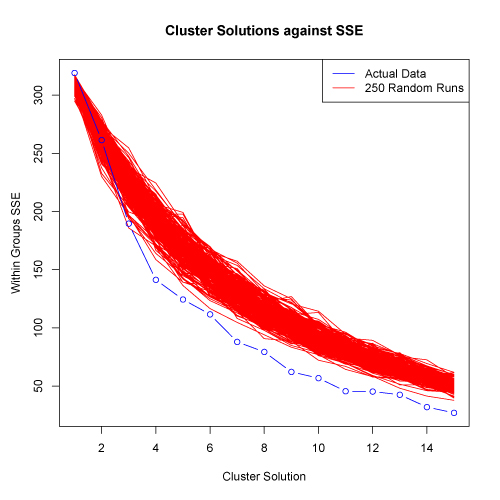
click to see images full size
In the examples shown above, the SSE for the actual data does decrease faster than the 250 randomized data sets. This suggests that the data set has structure and clusters are present. There is somewhat of a reduction in the rate of SSE decrease at about the 4 cluster solution. However, the "elbow" in the plot is not extreme and thus, further evaluation would be appropriate.
Another way to evaluate the appropriate cluster solution is to examine the absolute difference between the actual and random SSE against the tested cluster solutions. An appropriate cluster solution could be defined as the solution at which the actual SSE differs the most from the mean of the random SSE. To facilitate this comparison, the kmeans.R script displays the absolute difference between the actual and random (mean of all runs) SSE against the cluster solutions. One standard deviation above and below the mean absolute difference are also shown. Plots below are shown on both a log scale (left) and on a normal scale (right)
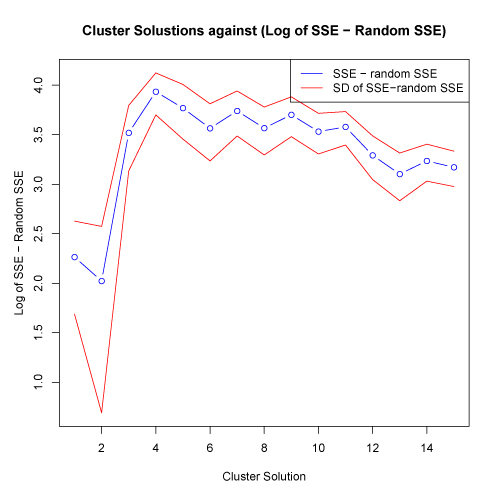
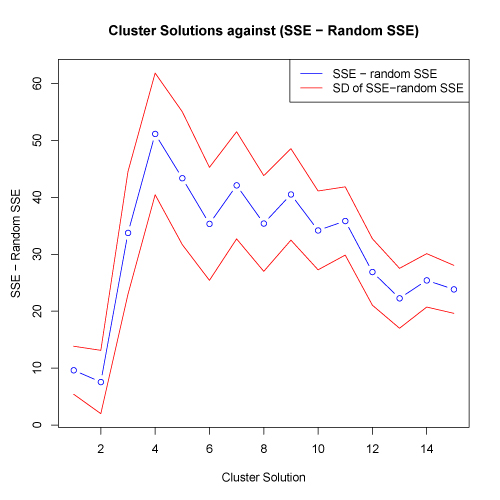
click to see images full size
In the plots above, the greatest absolute difference between actual and random SSE occurs at the 4 cluster solution. This suggests that this cluster solution may be an appropriate level to test. The fact that this cluster solution also coincides with the possible "elbow" in the plot of SSE against random SSE shown above provides additional information supporting this cluster solution.
The kmeans.R script provides one final plot that may sometimes be useful in evaluating the proper cluster solution. After the user selects a cluster solution, the script conducts a principal components analysis on the original data set. Each sample is then displayed on a scatter plot of the first two principal axes of the PCA with the clusters outlined. If the clusters are strong at the selected level, there should not be substantial overlap in the distributions of the cluster outlines on the PCA plot. It is important to note that PCA plots may not be particularly useful for K-means analysis of data sets with a large number of samples or a large number of variables. There is no hard and fast rule for this, but the percent of the variability explained by the PCA provides some clue as to the potential utility of this approach. Check here for more details. The examples provided below show a PCA plot of relatively strong clusters (left) and somewhat weaker clusters (right).
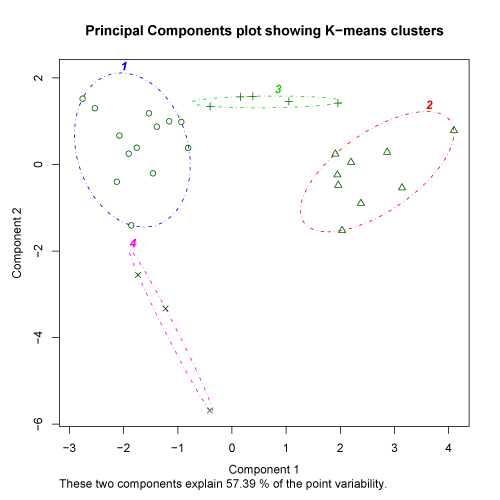
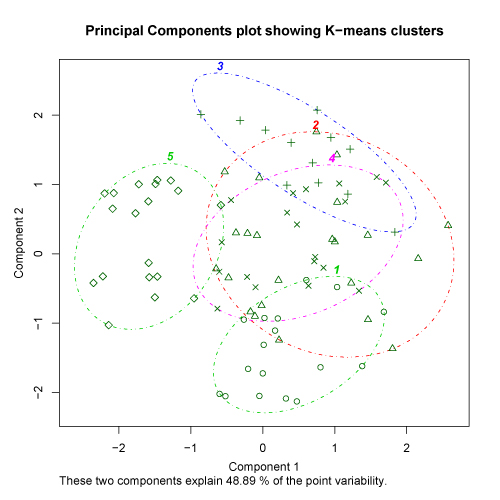
click to see images full size
It is important to note that all of the procedures described above are simply heuristics to aid in choosing the appropriate clustering level. None of the specific recommendations shown here should be interpreted as strict rules. It is preferable that these suggestions are used as general guidelines. In your own analysis, you should evaluate the results of each of these procedures against each other and against your own knowledge of the data set that you are using. For additional information on K-means cluster analysis, and its applications in archaeology, see the references below.
Additional References
B. S. Everitt, S. Landau and M. Leese
2001 Cluster Analysis. London, Edward Arnold.
Kintigh, Keith W.
1990 Intrasite Spatial Analysis: A Commentary on Major Methods. In Mathematics and Information Science in Archaeology: A Flexible Framework, edited by A. Voorrips, pp. 165-200. Studies in Modern Archaeology. vol. 3. HOLOS-Verlag, Bonn.
Kintigh, K. W., and A. J. Ammerman
1982 Heuristic Approaches to Spatial Analysis in Archaeology. American Antiquity 47:31-63.
Script: